PMMoV RNA Positive Control
PMMoV kit (100 reactions): A positive control designed to specifically for wastewater samples to detect PMMoV genomic RNA in a singleplex or multiplex reactions.
(RUO). Research Use Only. Not for use in Diagnostic Procedures.
Introduction
The DNA Software positive control PMMoV kit is intended to verify the validity of a real-time reverse transcriptase polymerase chain reaction (RT-PCR) on a sample of environmental origin, e.g., wastewater. This kit is for research use only and should not be used for diagnostic procedures.
The pepper mild mottle virus (PMMoV) is a plant (+)sense RNA virus that is highly abundant in human feces. It serves as a reliable internal control when detecting RNA from human pathogens by RT-PCR reaction.1, 2, 3
This kit is pre-validated with RNA extract from wastewater samples generously provided by Life Magnetics, Inc.

Tube 1: 20X primers/probe specific for PMMoV genomic RNA.
Contents
A mix of primers/probe targeting PMMoV genomic RNA is provided in a tube (a 20X concentrated working solution). The fluorophore of the probe is HEX™ (Hexachloro-fluorescein, a trademark of Life Technologies, Inc), and the quencher is BHQ-1™ (Black Hole Quencher, a trademark of Biosearch Technologies, Inc.).
Note: molecular biology grade water should be used to prepare the PCR reactions, which is NOT included in this kit.
Kit Handling and Contamination
The DNA Software PMMoV kit is shipped at room temperature, but for long-term it should be stored at -15°C or lower. The kit should be kept on ice once thawed.
Any contamination should be avoided by using appropriate personal protective equipment (PPE), powder free gloves, aerosol barrier pipette tips, and a clean hood.
Experimental
Set up your reaction (20 µL) as follows on ice:
| Component | Volume (µL) |
|---|---|
| TaqMan mastermix | 10 |
| RT enzyme | 0.5 |
| Target(s) primers/probe mix | 1 |
| PMMoV RNA primers/probe mix | 1 |
| Sample | 2 |
| Water | 5.5 |
Note: The volume of water should be adjusted accordingly if the user’s reaction preparation is different from the recommended preparation method.
An RT-PCR protocol was used in-house for pre-validation on a Bio-Rad CFX96™ Real-Time System, with the following program:
| Step | Thermocycling Protocol |
|---|---|
| 1 | Incubate @ 50 °C for 5 minutes |
| 2 | Incubate @ 94 °C for 3 minutes |
| 3 | Incubate @ 94 °C for 5 seconds |
| 4 | Incubate @ 63 °C for 30 seconds |
| 5 | Plate Read |
| 6 | Go to Step 3, repeat 44x more |
| 7 | (optional) Incubate @ 63 °C for 3 minutes |
Result Interpretation
After running the qPCR reaction, perform a regression analysis on the data to determine the quantification cycle, Cq. (Cq is preferred over Ct). Each fluorescence channel with a Cq < 38 cycles and final RFU >200 is considered “positive” or “+” in the Table below.
| Target RNA (Fluorophore) | PMMoV (HEX™) | Recommended Interpretation |
|---|---|---|
| – | – | The PCR reaction failed. Please repeat the experiment |
| – | + | The sample doesn’t contain the target RNA. |
| + | – | The sample contains the target RNA. The sample may not contain PMMoV RNA. |
| + | + | The sample contains the target RNA, and PMMoV RNA. |
Pre-Validation
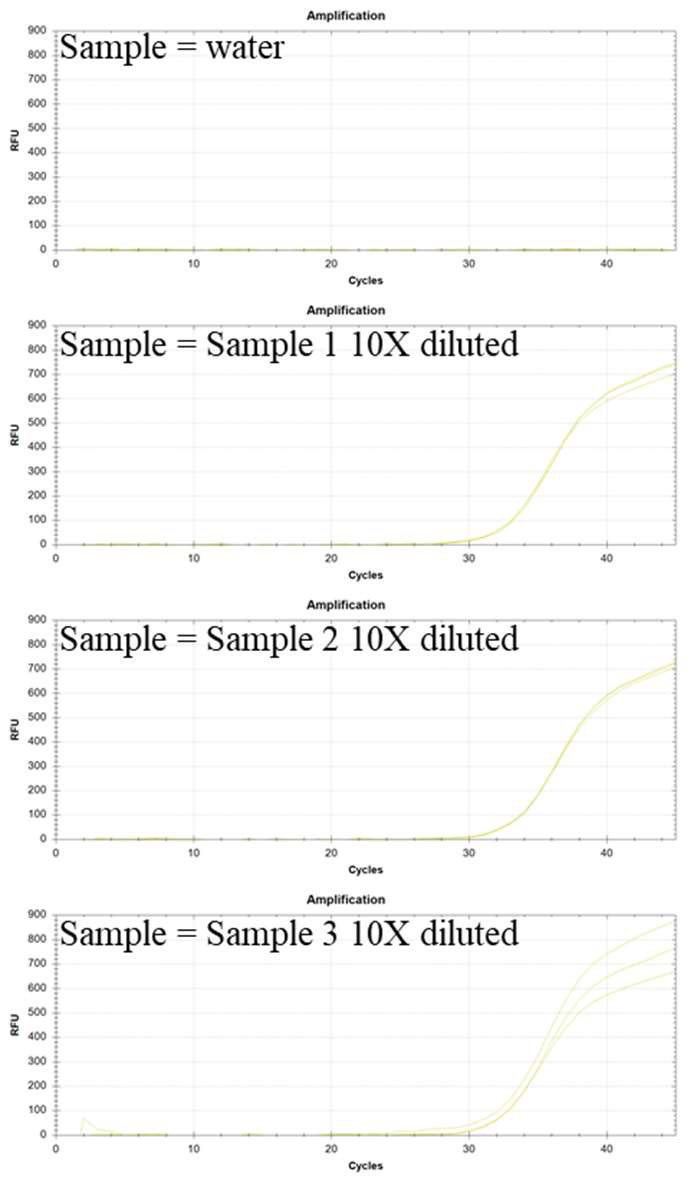
Experiments were performed in triplicate using a protocol modified from the experimental procedure given above. Only primers and probe from the PMMoV RNA kit were employed, without any primers or probe for other targets. The samples used for the validation experiments included water (NTC) and three 10X diluted RNA extract solution from local wastewater samples. The reverse transcriptase and TaqMan mastermix from Empirical Bioscience (items: RTqPCR-Kit-200) were employed. The results of these experiments are shown in Figure 1 below:
Figure 1: Validation experiments with water (no reaction observed) and three RNA extract samples from wastewater from three different locations in Michigan (for PCR each sample was 10X diluted). The HEX probe detects PMMoV genomic RNA.
Conclusion: The data in Figure 1 indicates that the PMMoV kit can detect PMMoV genomic RNA in wastewater samples.
Contact Us
For assistance, please contact DNA Software using the link: https://www.dnasoftware.com/contact/
Address:
Michigan Life Science and Innovation Center
46701 Commerce Center Dr
Plymouth, MI 48170
Phone: (734) 222-9080
Literature Reference
- 1. D’Aoust PM, Mercier E, Montpetit D, et al. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. (2021) 188, 116560. 2.
- Rosario, K., Symonds, E.M., Sinigalliano, C., et al. Pepper Mild Mottle Virus as an Indicator of Fecal Pollution. Appl. Environ. Microbio. (2009) 75 (22), 7261. 3.
- Ahmed W., Bivins, A., Bertsch, P.M., et al. Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimization and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci, Health.(2020) 17, 100209.